Demo Inference Service#
Demo notebook for sending a prediction request to the deployed inference service, and visualizing the results.
Requirements#
! pip install numpy==1.26.4 einops==0.4.1 scipy==1.10.1 connected-components-3d monai[nibabel,pillow,ignite,tqdm,pydicom]====1.5.0 synapseclient
Import utility functions#
from utils import plot_input_scan, send_prediction_request, plot_results, plot_results_overlap
Dataset#
Download scan image from the Multi-Atlas Labeling Beyond the Cranial Vault - Workshop and Challenge dataset.
You must create a Synapse account and get Personal Access Token (PAT) to access the dataset.
pip install synapseclient
synapse get syn3553734
unzip Abdomen.zip
The data contains labels for 13 different organs. Check this link for additional details on the dataset: Abdomen dataset.
(1) spleen
(2) right kidney
(3) left kidney
(4) gallbladder
(5) esophagus
(6) liver
(7) stomach
(8) aorta
(9) inferior vena cava
(10) portal vein and splenic vein
(11) pancreas
(12) right adrenal gland
(13) left adrenal gland
Port forwarding to inference service#
Open your terminal and run the following command:
kubectl port-forward -n <namespace> service/dev-lifescience-swinunetr-<release-name> 8000:80Keep the Terminal Open: The port-forward command runs in the foreground. You must keep this terminal window open for the connection to remain active.
Inference Parameters#
# Path to the image file to send for prediction.
image_path = "./Abdomen/RawData/Training/img/img0001.nii.gz"
# Ground truth label file for the image.
label_path = "./Abdomen/RawData/Training/label/label0001.nii.gz"
# Filename to save the output prediction
output_path = "client_prediction.nii.gz"
# URL of the prediction inference service endpoint
service_url = "http://localhost:8000/predict/"
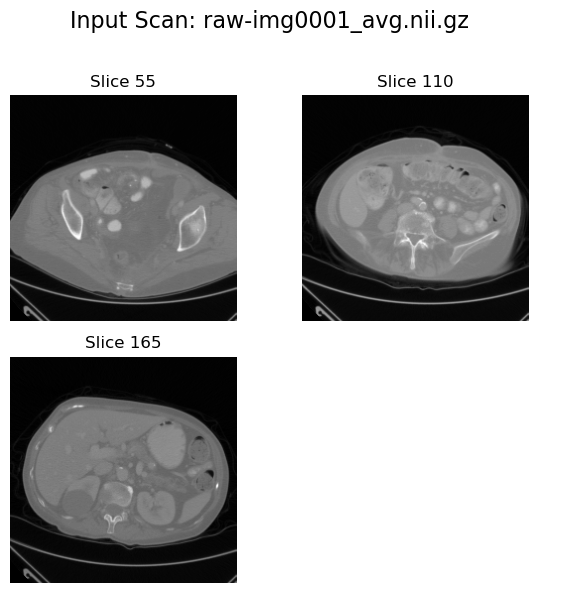
Input view#
plot_input_scan(input_path=image_path, num_slices_to_plot=3)
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:51:45,765 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:51:49,784 - INFO - Input data shape: torch.Size([229, 229, 220])
Send prediction request#
Send a CT san to the inference service for prediction:
send_prediction_request(image_path=image_path, output_path=output_path, server_url=service_url)
INFO:utils:Attempting to send ../../data/inputs_nifti/LUNG1-001_input.nii.gz to http://localhost:8001/predict/
INFO:utils:Status Code: 200
INFO:utils:Success! Prediction saved to client_prediction.nii.gz
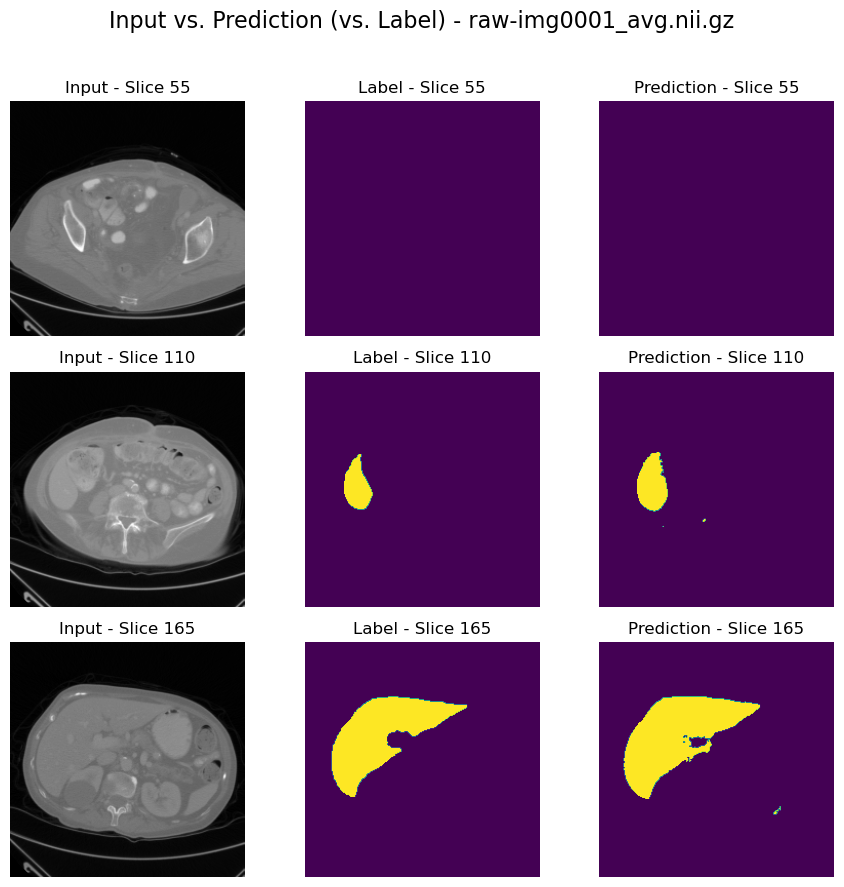
Visualize results#
channel_idx = 6
plot_results(image_path, label_path, output_path, channel_idx=channel_idx, num_slices_to_plot=3)
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:52:08,516 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:52:10,516 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
Input data shape: torch.Size([1, 229, 229, 220])
Label data shape: torch.Size([14, 229, 229, 220])
Prediction data shape: torch.Size([14, 229, 229, 220])
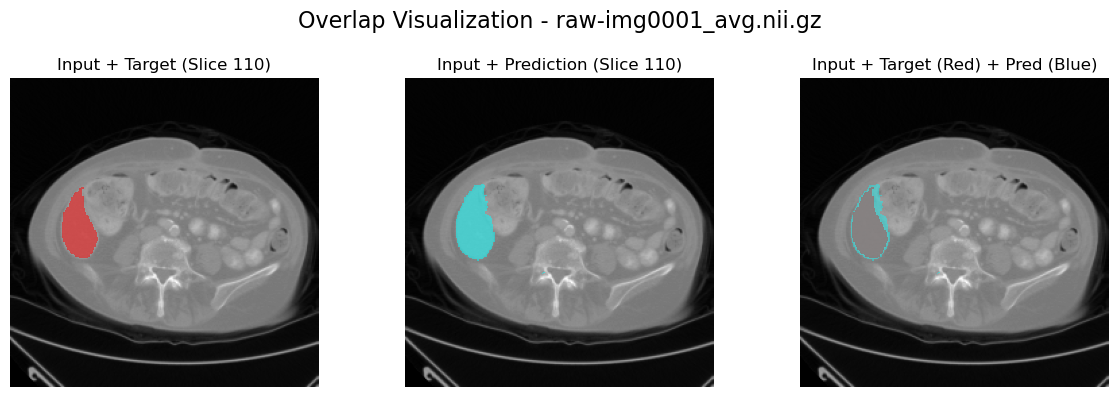
slice_to_plot = 110
plot_results_overlap(
image_path, label_path, output_path, channel_idx=channel_idx, slice_to_plot=slice_to_plot
)
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:53:00,148 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:53:02,367 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
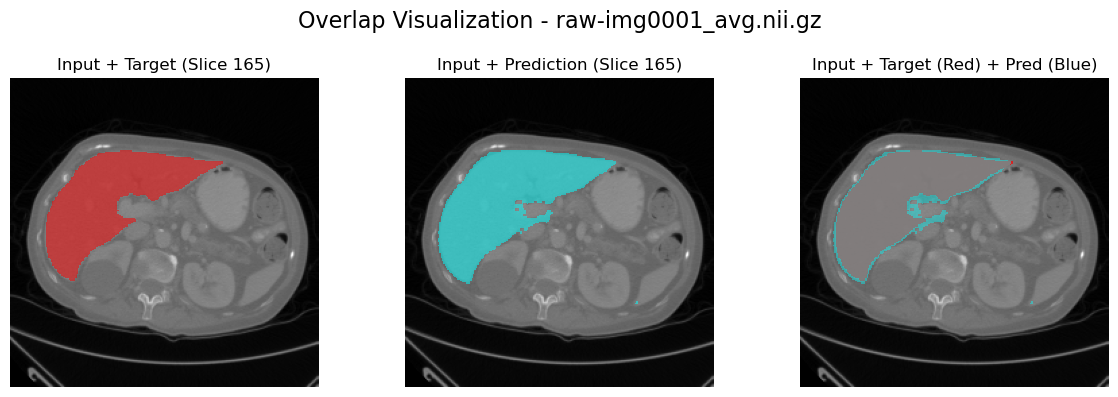
channel_idx = 6
slice_to_plot = 165
plot_results_overlap(
image_path, label_path, output_path, channel_idx=channel_idx, slice_to_plot=slice_to_plot
)
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:53:42,010 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1
2025-08-04 10:53:44,120 - INFO - pixdim[0] (qfac) should be 1 (default) or -1; setting qfac to 1